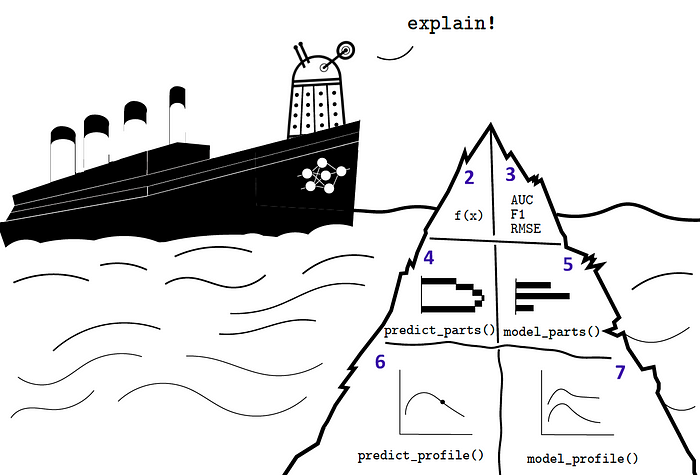
Explainability
Explanatory Model Analysis Book with examples in Python
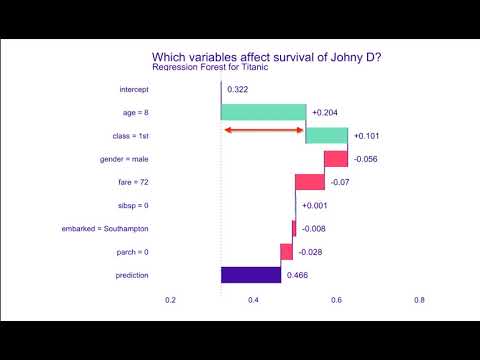
Introduction to dalex Titanic: tutorial and examples
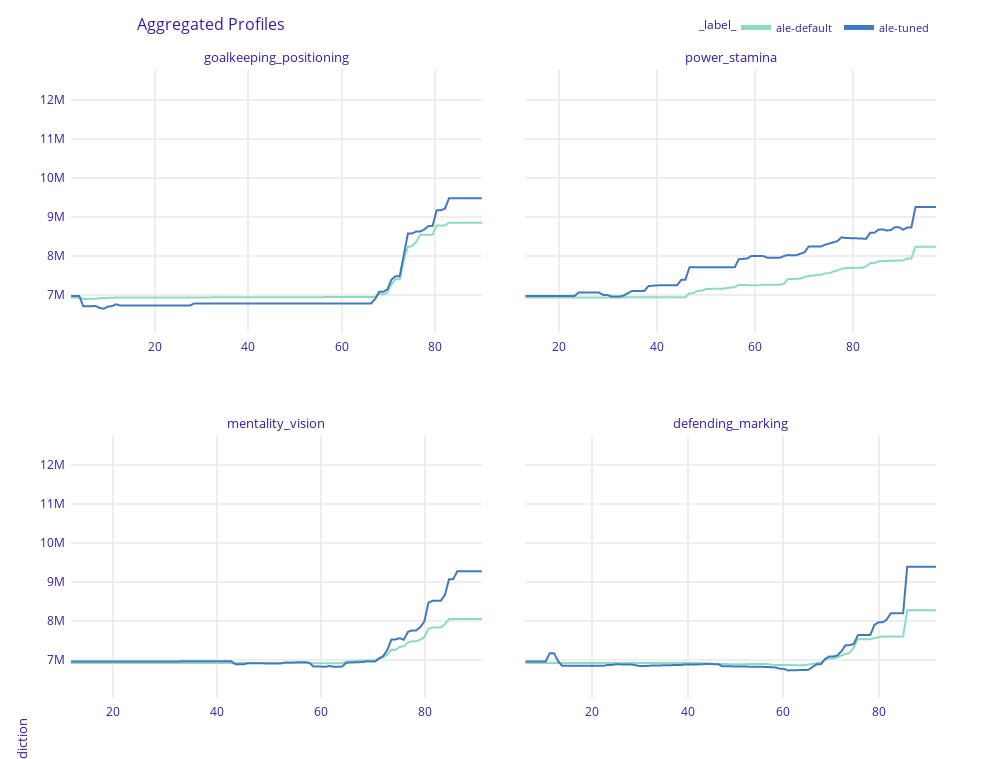
Key features explained FIFA 20: explain default vs tuned model with dalex

Multioutput predictive models Explaining multiclass classification and multioutput regression
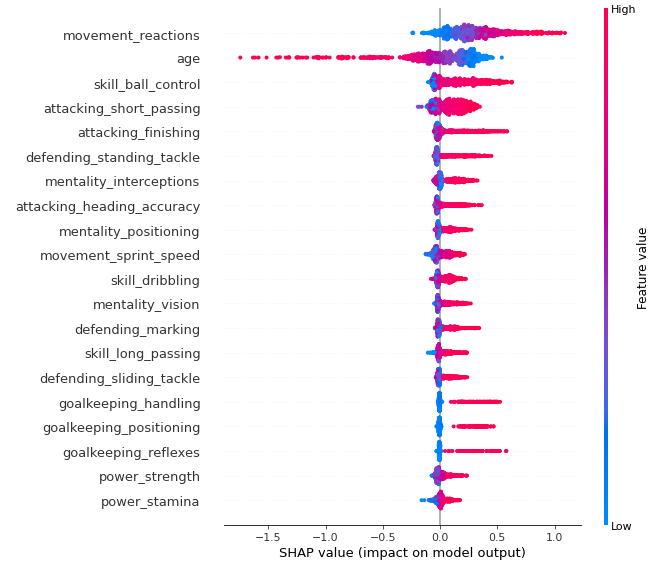
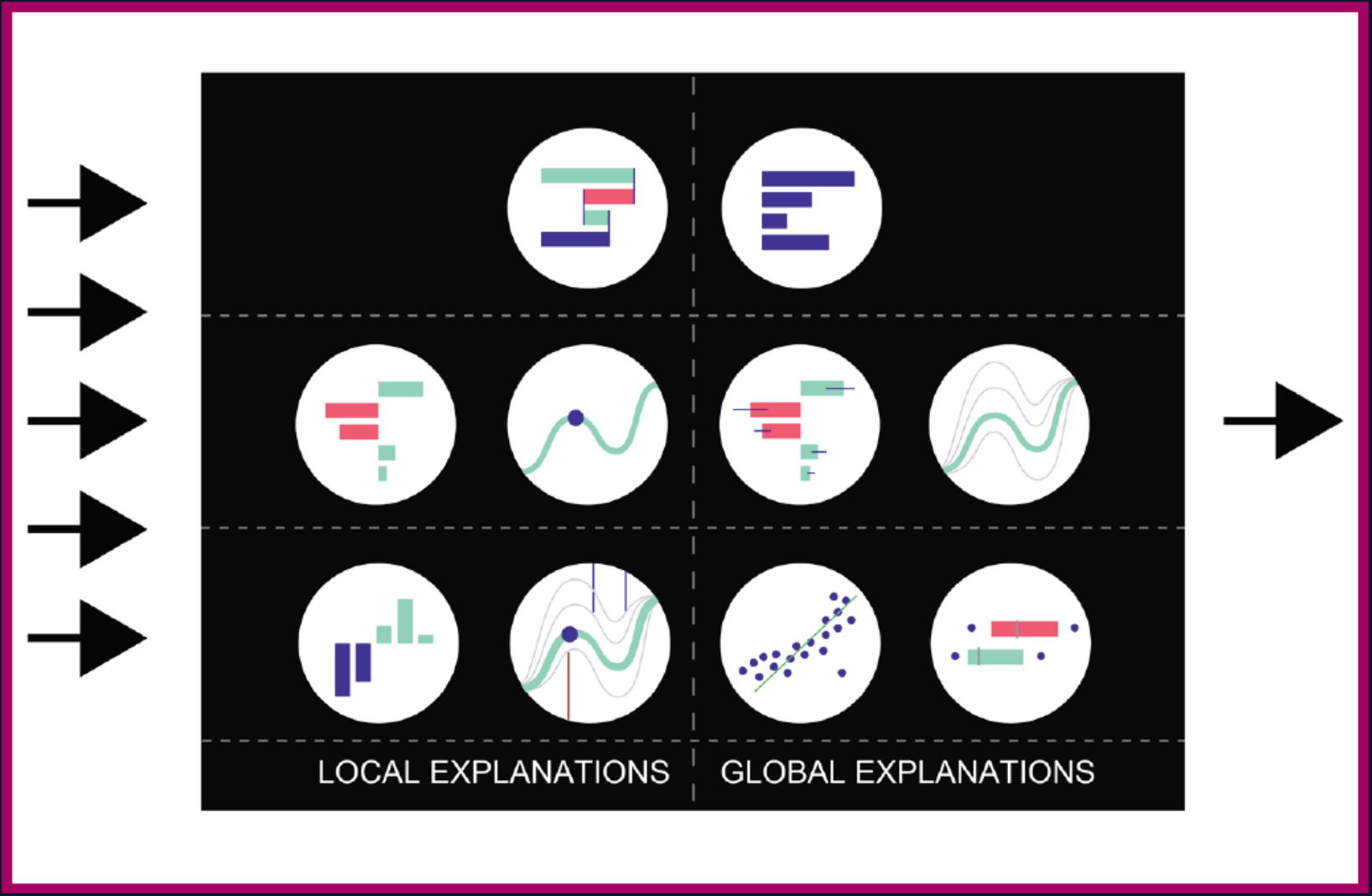
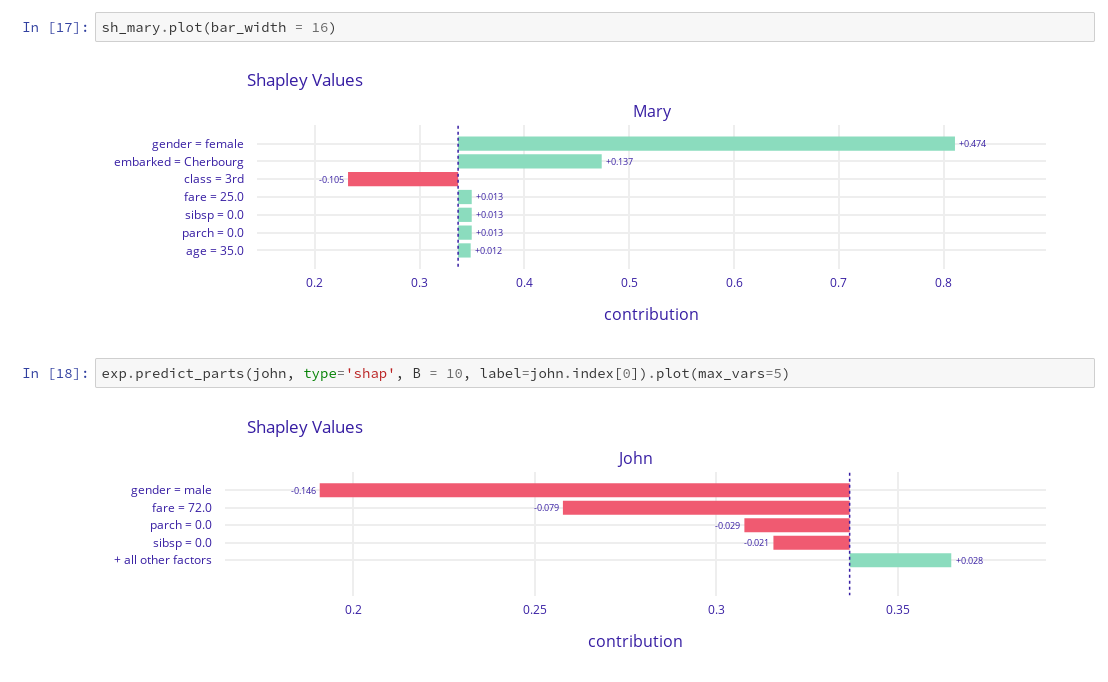
More explanations residuals, shap, lime
Tutorial: Basic XAI
Fairness
Interactive analysis
Materials & blogs
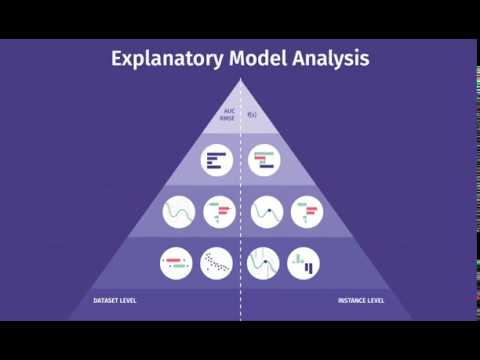
Introductory videos
Documentation

API Reference Python documentation with pdoc

Paper Journal of Machine Learning Research
Citation BibTeX format
GitHub repository with code
How to add a new model? Developer instruction: model

How to add a new explanation? Developer instruction: explanation

How to add a new fairness metric? Developer instruction: fairness metric
Attributions
TensorFlow, the TensorFlow logo and any related marks are trademarks of Google Inc.
XGBoost logo are treademarks of Distributed (Deep) Machine Learning Community.