Titanic: tutorial and examples¶
imports¶
import dalex as dx
import pandas as pd
import numpy as np
from sklearn.neural_network import MLPClassifier
from sklearn.preprocessing import StandardScaler, OneHotEncoder
from sklearn.impute import SimpleImputer
from sklearn.pipeline import Pipeline
from sklearn.compose import ColumnTransformer
import warnings
warnings.filterwarnings('ignore')
import plotly
plotly.offline.init_notebook_mode()
dx.__version__
load data¶
First, divide the data into variables X and a target variable y.
data = dx.datasets.load_titanic()
X = data.drop(columns='survived')
y = data.survived
data.head(10)
create a pipeline model¶
numerical_transformerpipeline:numerical_features: choose numerical features to transform- impute missing data with median strategy
- scale numerical features with standard scaler
categorical_transformerpipeline:categorical_features: choose categorical features to transform- impute missing data with
'missing'string - encode categorical features with one-hot
aggregate those two pipelines into a
preprocessorusingColumnTransformermake a basic
classifiermodel usingMLPClassifier- it has 3 hidden layers with sizes 150, 100, 50 respectivelyconstruct a
clfpipeline model, which combines thepreprocessorwith the basicclassifiermodel
numerical_features = ['age', 'fare', 'sibsp', 'parch']
numerical_transformer = Pipeline(
steps=[
('imputer', SimpleImputer(strategy='median')),
('scaler', StandardScaler())
]
)
categorical_features = ['gender', 'class', 'embarked']
categorical_transformer = Pipeline(
steps=[
('imputer', SimpleImputer(strategy='constant', fill_value='missing')),
('onehot', OneHotEncoder(handle_unknown='ignore'))
]
)
preprocessor = ColumnTransformer(
transformers=[
('num', numerical_transformer, numerical_features),
('cat', categorical_transformer, categorical_features)
]
)
classifier = MLPClassifier(hidden_layer_sizes=(150,100,50), max_iter=500, random_state=0)
clf = Pipeline(steps=[('preprocessor', preprocessor),
('classifier', classifier)])
fit the model¶
clf.fit(X, y)
create an explainer for the model¶
exp = dx.Explainer(clf, X, y)
introduction to the topic: Explanatory Model Analysis: Explore, Explain, and Examine Predictive Models¶
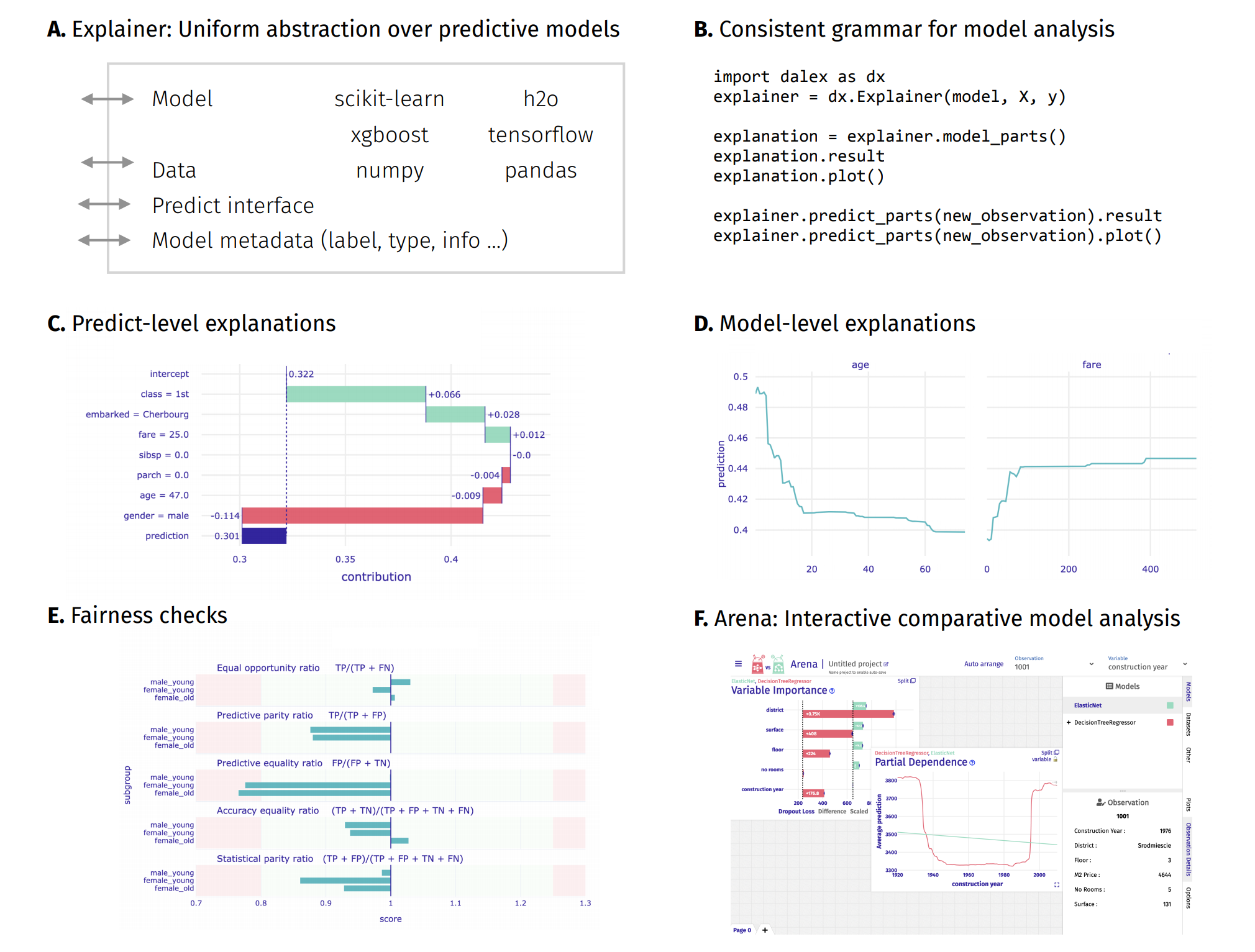
Above functionalities are accessible from the Explainer object through its methods.
Model-level and predict-level methods return a new unique object that contains the result attribute (pandas.DataFrame) and the plot method.
predict¶
This function is nothing but normal model prediction, however it uses Explainer interface.
Let's create two example persons for this tutorial.
john = pd.DataFrame({'gender': ['male'],
'age': [25],
'class': ['1st'],
'embarked': ['Southampton'],
'fare': [72],
'sibsp': [0],
'parch': 0},
index = ['John'])
mary = pd.DataFrame({'gender': ['female'],
'age': [35],
'class': ['3rd'],
'embarked': ['Cherbourg'],
'fare': [25],
'sibsp': [0],
'parch': [0]},
index = ['Mary'])
You can make a prediction on many samples at the same time
exp.predict(X)[0:10]
As well as on only one instance. However, the only accepted format is pandas.DataFrame.
Prediction of survival for John.
exp.predict(john)
Prediction of survival for Mary.
exp.predict(mary)
predict_parts¶
'break_down''break_down_interactions''shap'
This function calculates Variable Attributions as Break Down, iBreakDown or Shapley Values explanations.
Model prediction is decomposed into parts that are attributed for particular variables.
bd_john = exp.predict_parts(john, type='break_down', label=john.index[0])
bd_interactions_john = exp.predict_parts(john, type='break_down_interactions', label="John+")
sh_mary = exp.predict_parts(mary, type='shap', B=10, label=mary.index[0])
bd_john.result
bd_john.plot(bd_interactions_john)
sh_mary.result.loc[sh_mary.result.B == 0, ]
sh_mary.plot(bar_width=16)
exp.predict_parts(john, type='shap', B=10, label=john.index[0]).plot(max_vars=5)
predict_profile¶
'ceteris_paribus'
This function computes individual profiles aka Ceteris Paribus Profiles.
cp_mary = exp.predict_profile(mary, label=mary.index[0])
cp_john = exp.predict_profile(john, label=john.index[0])
cp_mary.result.head()
cp_mary.plot(cp_john)
cp_john.plot(cp_mary, variable_type = "categorical")
model_performance¶
'classification''regression'
This function calculates various Model Performance measures:
- classification: F1, accuracy, recall, precision and AUC
- regression: mean squared error, R squared, median absolute deviation
mp = exp.model_performance(model_type = 'classification')
mp.result
mp.result.auc[0]
mp.plot(geom="roc")
model_parts¶
'variable_importance''ratio''difference'
This function calculates Variable Importance.
vi = exp.model_parts()
vi.result
vi.plot(max_vars=5)
There is also a possibility of calculating variable importance of group of variables.
vi_grouped = exp.model_parts(variable_groups={'personal': ['gender', 'age', 'sibsp', 'parch'],
'wealth': ['class', 'fare']})
vi_grouped.result
vi_grouped.plot()
model_profile¶
'partial''accumulated'
This function calculates explanations that explore model response as a function of selected variables.
The explanations can be calulated as Partial Dependence Profile or Accumulated Local Dependence Profile.
pdp_num = exp.model_profile(type = 'partial', label="pdp")
ale_num = exp.model_profile(type = 'accumulated', label="ale")
pdp_num.plot(ale_num)
pdp_cat = exp.model_profile(type = 'partial', variable_type='categorical',
variables = ["gender","class"], label="pdp")
ale_cat = exp.model_profile(type = 'accumulated', variable_type='categorical',
variables = ["gender","class"], label="ale")
ale_cat.plot(pdp_cat)
Hover over all of the above plots for tooltips with more information.
Saving and loading Explainers¶
You can easily save an Explainer to the pickle (or generaly binary form) and load it again. Any local or lambda function in the Explainer object will be dropped during saving. Residual function by default is local, thus, if default, it is always dropped. Default functions can be retrieved during loading.
# this converts explainer to a binary form
# exp.dumps()
# this load explainer again
# dx.Explainer.loads(pickled)
# this will not retrieve default function if dropped
# dx.Explainer.loads(pickled, use_defaults=False)
# this will save your explainer to the file
# with open('explainer.pkl', 'wb') as fd:
# exp.dump(fd)
# this will load your explainer from the file
# with open('explainer.pkl', 'rb') as fd:
# dx.Explainer.load(fd)
Plots¶
This package uses plotly to render the plots:
- Install extentions to use
plotlyin JupyterLab: Getting Started Troubleshooting - Use
show=Falseparameter inplotmethod to returnplotly Figureobject - It is possible to edit the figures and save them
Resources - https://dalex.drwhy.ai/python¶
Introduction to the
dalexpackage: Titanic: tutorial and examplesKey features explained: FIFA20: explain default vs tuned model with dalex
How to use dalex with: xgboost, tensorflow, h2o (feat. autokeras, catboost, lightgbm)
More explanations: residuals, shap, lime
Introduction to the Fairness module in dalex
Introduction to the Aspect module in dalex
Introduction to Arena: interactive dashboard for model exploration
Code in the form of jupyter notebook
Changelog: NEWS
Theoretical introduction to the plots: Explanatory Model Analysis: Explore, Explain, and Examine Predictive Models